Bootstrap CIs for Standardized Solution
Source:R/standardizedSolution_boot_ci.R
standardizedSolution_boot.RdFunctions for forming bootstrap confidence intervals for the standardized solution.
Usage
standardizedSolution_boot(
object,
level = 0.95,
type = "std.all",
boot_delta_ratio = FALSE,
boot_ci_type = c("perc", "bc", "bca.simple"),
save_boot_est_std = TRUE,
boot_pvalue = TRUE,
boot_pvalue_min_size = 1000,
...
)Arguments
- object
A 'lavaan'-class object, fitted with 'se = "boot"'.
- level
The level of confidence of the confidence intervals. Default is .95.
- type
The type of standard estimates. The same argument of
lavaan::standardizedSolution(), and support all values supported bylavaan::standardizedSolution(). Default is"std.all".- boot_delta_ratio
The ratio of (a) the distance of the bootstrap confidence limit from the point estimate to (b) the distance of the delta-method limit from the point estimate. Default is
FALSE.- boot_ci_type
The type of the bootstrapping confidence intervals. Support percentile confidence intervals (
"perc", the default) and bias-corrected confidence intervals ("bc"or"bca.simple").- save_boot_est_std
Whether the bootstrap estimates of the standardized solution are saved. If saved, they will be stored in the attribute
boot_est_std. Default isTRUE.- boot_pvalue
Whether asymmetric bootstrap p-values are computed. Default is
TRUE.- boot_pvalue_min_size
Integer. The asymmetric bootstrap p-values will be computed only if the number of valid bootstrap estimates is at least this value. Otherwise,
NAwill be returned. If the number of valid bootstrap samples is less than this value, thenboot_pvaluewill be set toFALSE.- ...
Other arguments to be passed to
lavaan::standardizedSolution().
Value
The output of
lavaan::standardizedSolution(),
with bootstrap confidence intervals
appended to the right, with class
set to sbt_std_boot. It has
a print method
(print.sbt_std_boot()) that
can be used to print the standardized
solution in a format similar to
that of the printout of
the summary() of a lavaan::lavaan object.
Details
standardizedSolution_boot()
receives a
lavaan::lavaan object fitted
with bootstrapping standard errors
requested and forms the confidence
intervals for the standardized
solution.
It works by calling
lavaan::standardizedSolution()
with the bootstrap estimates
of free parameters in each bootstrap sample
to compute the standardized estimates
in each sample.
Alternative, call store_boot() to
computes and store bootstrap estimates
of the standardized solution.
This function will then retrieve them,
even if se was not set to
"boot" or "bootstrap" when fitting
the model.
Bootstrap Confidence Intervals
It supports percentile and bias-corrected bootstrap confidence intervals.
References
Asparouhov, A., & Muthén, B. (2021). Bootstrap p-value computation. Retrieved from https://www.statmodel.com/download/FAQ-Bootstrap%20-%20Pvalue.pdf
Author
Shu Fai Cheung
https://orcid.org/0000-0002-9871-9448.
Originally proposed in an issue at GitHub
https://github.com/simsem/semTools/issues/101#issue-1021974657,
inspired by a discussion at
the Google group for lavaan
https://groups.google.com/g/lavaan/c/qQBXSz5cd0o/m/R8YT5HxNAgAJ.
boot::boot.ci() is used to form the
percentile confidence intervals in
this version.
Examples
library(lavaan)
set.seed(5478374)
n <- 50
x <- runif(n) - .5
m <- .40 * x + rnorm(n, 0, sqrt(1 - .40))
y <- .30 * m + rnorm(n, 0, sqrt(1 - .30))
dat <- data.frame(x = x, y = y, m = m)
model <-
'
m ~ a*x
y ~ b*m
ab := a*b
'
# Should set bootstrap to at least 2000 in real studies
fit <- sem(model, data = dat, fixed.x = FALSE,
se = "boot",
bootstrap = 100)
summary(fit)
#> lavaan 0.6-21 ended normally after 1 iteration
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of model parameters 5
#>
#> Number of observations 50
#>
#> Model Test User Model:
#>
#> Test statistic 0.020
#> Degrees of freedom 1
#> P-value (Chi-square) 0.887
#>
#> Parameter Estimates:
#>
#> Standard errors Bootstrap
#> Number of requested bootstrap draws 100
#> Number of successful bootstrap draws 100
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|)
#> m ~
#> x (a) 0.569 0.325 1.749 0.080
#> y ~
#> m (b) 0.219 0.146 1.495 0.135
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|)
#> .m 0.460 0.086 5.381 0.000
#> .y 0.570 0.110 5.178 0.000
#> x 0.078 0.012 6.782 0.000
#>
#> Defined Parameters:
#> Estimate Std.Err z-value P(>|z|)
#> ab 0.125 0.125 0.997 0.319
#>
std <- standardizedSolution_boot(fit)
#> Warning: The number of bootstrap samples (100) is less than 'boot_pvalue_min_size' (1000). Bootstrap p-values are not computed.
std
#>
#> Bootstrapping:
#>
#> Valid Bootstrap Samples: 100
#> Level of Confidence: 95.0%
#> CI Type: Percentile
#> Standardization Type: std.all
#>
#> Parameter Estimates Settings:
#>
#> Standard errors: Bootstrap
#> Number of requested bootstrap draws: 100
#> Number of successful bootstrap draws: 100
#>
#> Regressions:
#> Std SE p CI.Lo CI.Up bSE bCI.Lo bCI.Up
#> m ~
#> x (a) 0.229 0.127 0.072 -0.020 0.477 0.125 -0.041 0.454
#> y ~
#> m (b) 0.198 0.118 0.092 -0.032 0.429 0.115 -0.024 0.464
#>
#> Variances:
#> Std SE p CI.Lo CI.Up bSE bCI.Lo bCI.Up
#> .m 0.948 0.058 0.000 0.834 1.062 0.057 0.793 1.000
#> .y 0.961 0.047 0.000 0.869 1.052 0.052 0.785 1.000
#> x 1.000
#>
#> Defined Parameters:
#> Std SE p CI.Lo CI.Up bSE bCI.Lo bCI.Up
#> ab (ab) 0.045 0.040 0.259 -0.033 0.124 0.043 -0.007 0.164
#>
#> Footnote:
#> - Std: Standardized estimates.
#> - SE: Delta method standard errors.
#> - p: Delta method p-values.
#> - CI.Lo, CI.Up: Delta method confidence intervals.
#> - bSE: Bootstrap standard errors.
#> - bCI.Lo, bCI.Up: Bootstrap confidence intervals.
# Print in a friendly format with only standardized solution
print(std, output = "text")
#>
#> Standardized Estimates Only
#>
#> Standard errors Bootstrap
#> Standard errors (boot.se) Bootstrap
#> Confidence interval (boot.ci.) Bootstrap
#> Confidence Level (boot.ci.) 95.0%
#> Bootstrap CI Type (boot.ci.) Percentile
#> Standardization Type std.all
#> Number of requested bootstrap draws 100
#> Number of successful bootstrap draws 100
#>
#> Regressions:
#> Standardized Std.Err z-value P(>|z|) ci.lower ci.upper
#> m ~
#> x (a) 0.229 0.127 1.800 0.072 -0.020 0.477
#> y ~
#> m (b) 0.198 0.118 1.684 0.092 -0.032 0.429
#> boot.se boot.ci.lower boot.ci.upper
#>
#> 0.125 -0.041 0.454
#>
#> 0.115 -0.024 0.464
#>
#> Variances:
#> Standardized Std.Err z-value P(>|z|) ci.lower ci.upper
#> .m 0.948 0.058 16.325 0.000 0.834 1.062
#> .y 0.961 0.047 20.595 0.000 0.869 1.052
#> x 1.000 1.000 1.000
#> boot.se boot.ci.lower boot.ci.upper
#> 0.057 0.793 1.000
#> 0.052 0.785 1.000
#> NA NA NA
#>
#> Defined Parameters:
#> Standardized Std.Err z-value P(>|z|) ci.lower ci.upper
#> ab 0.045 0.040 1.130 0.259 -0.033 0.124
#> boot.se boot.ci.lower boot.ci.upper
#> 0.043 -0.007 0.164
#>
# Print in a friendly format with both unstandardized
# and standardized solution
print(std, output = "text", standardized_only = FALSE)
#>
#> Parameter Estimates:
#>
#> Standard errors Bootstrap
#> Number of requested bootstrap draws 100
#> Number of successful bootstrap draws 100
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) ci.lower ci.upper
#> m ~
#> x (a) 0.569 0.325 1.749 0.080 -0.098 1.261
#> y ~
#> m (b) 0.219 0.146 1.495 0.135 -0.020 0.613
#> Standardized Std.Err.std ci.std.lower ci.std.upper
#>
#> 0.229 0.125 -0.041 0.454
#>
#> 0.198 0.115 -0.024 0.464
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) ci.lower ci.upper
#> .m 0.460 0.086 5.381 0.000 0.279 0.623
#> .y 0.570 0.110 5.178 0.000 0.345 0.775
#> x 0.078 0.012 6.782 0.000 0.055 0.101
#> Standardized Std.Err.std ci.std.lower ci.std.upper
#> 0.948 0.057 0.793 1.000
#> 0.961 0.052 0.785 1.000
#> 1.000 NA NA NA
#>
#> Defined Parameters:
#> Estimate Std.Err z-value P(>|z|) ci.lower ci.upper
#> ab 0.125 0.125 0.997 0.319 -0.019 0.501
#> Standardized Std.Err.std ci.std.lower ci.std.upper
#> 0.045 0.043 -0.007 0.164
#>
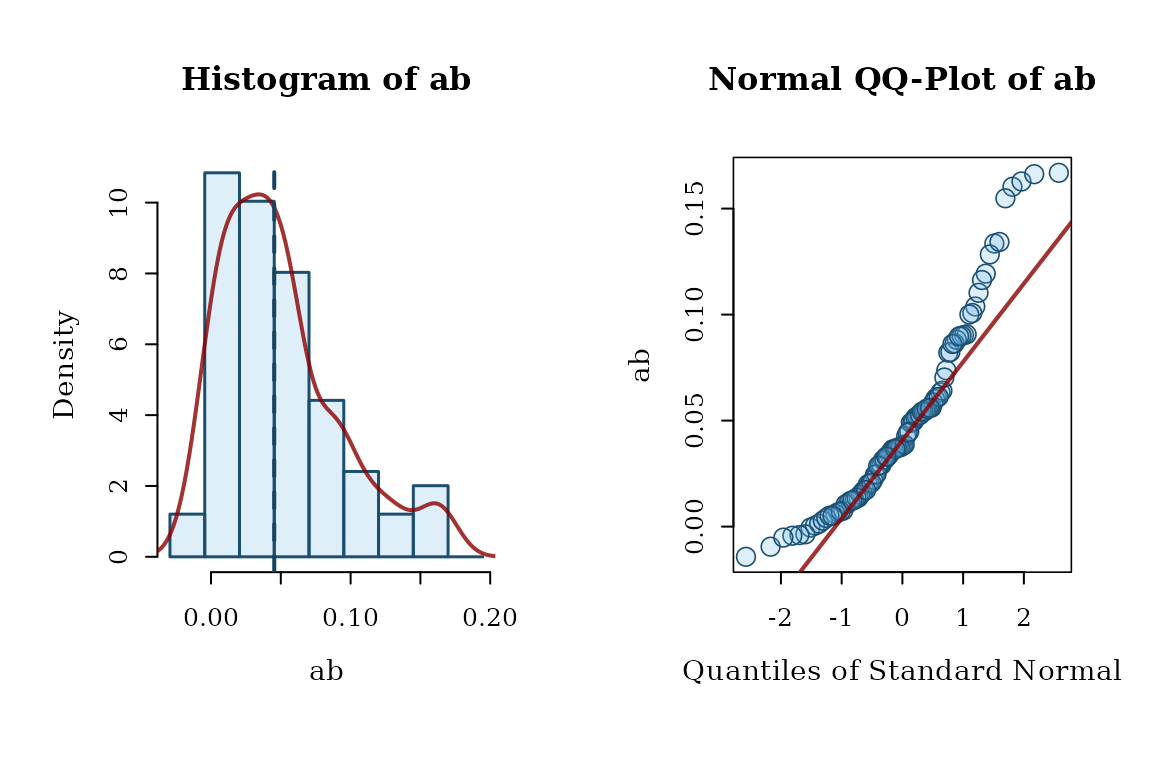
# hist_qq_boot() can be used to examine the bootstrap estimates
# of a parameter
hist_qq_boot(std, param = "ab")
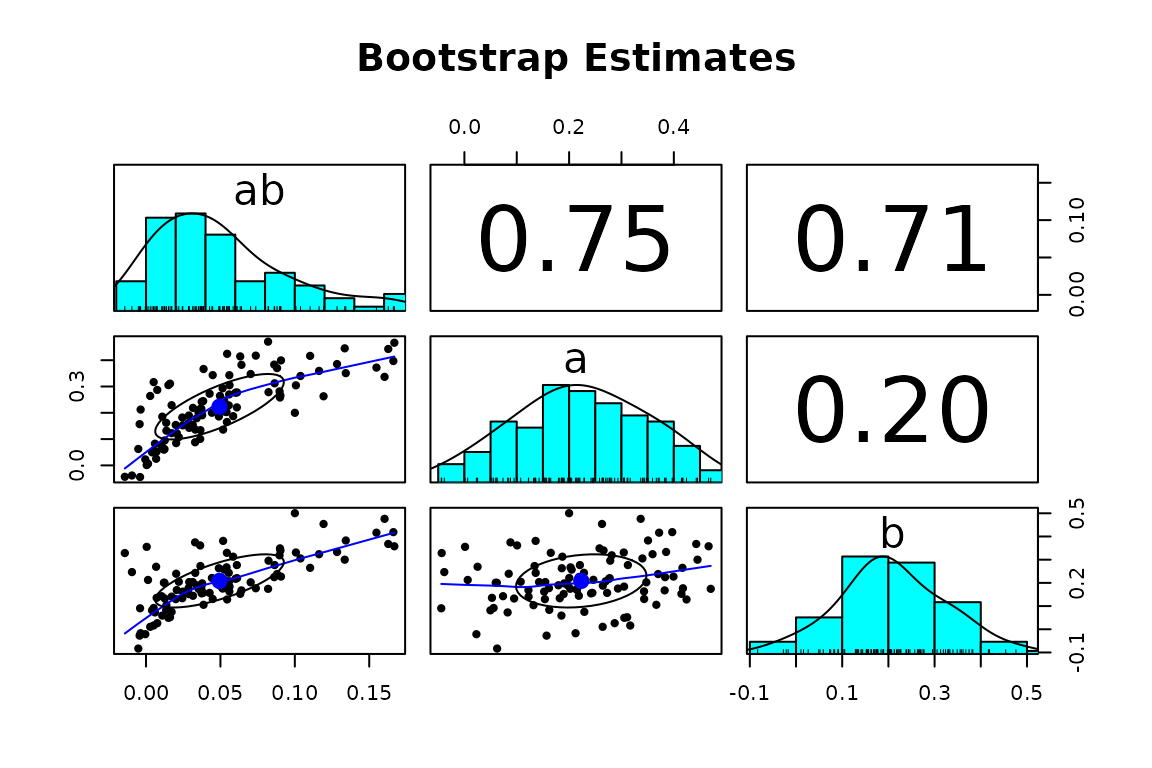
 # scatter_boot() can be used to examine the bootstrap estimates
# of two or more parameters
scatter_boot(std, params = c("ab", "a", "b"))
# scatter_boot() can be used to examine the bootstrap estimates
# of two or more parameters
scatter_boot(std, params = c("ab", "a", "b"))